Abstract
The effect and underlying mechanism of vitamin A on norovirus infection are largely unknown. This study aimed to investigate how vitamin A administration affects the gut microbiome after norovirus infection. Here, we demonstrate that treatment with either retinol or retinoic acid (RA) inhibits murine norovirus (MNV) replication using both in vitro and in vivo models. Compositional changes in the gut microbiome associated with RA administration and/or norovirus infection were also investigated. Oral administration of RA and/or MNV significantly altered intestinal microbiome profiles. Particularly, bacterial species belonging to the Lactobacillaceae families were remarkably increased by MNV inoculation and RA administration, suggesting that the antiviral effects of RA occur via the modulation of specific microbiota. The antiviral causal effect of Lactobacillus was identified and demonstrated using in vitro models in RAW264.7 cells. The antiviral immune response to MNV was mediated by IFN-β upregulation. This study represents the first comprehensive profiling of gut microbiota in response to RA treatment against norovirus infection. Moreover, we conclude that the abundance of Lactobacillus through gut microbiota modulation by RA is at least partially responsible for norovirus inhibition.
Similar content being viewed by others
Introduction
Norovirus is the most frequent etiological viral agent of acute gastroenteritis among all age groups worldwide. Norovirus causes approximately 90% of all epidemic nonbacterial outbreaks of gastroenteritis around the world, and is responsible for approximately 50% of all foodborne outbreaks of gastroenteritis in the United States and many other countries1,2. Viral infection causes various clinical symptoms, including diarrhea, vomiting, nausea, abdominal pain, and fever lasting one to three days. Unfortunately, there is no current treatment or vaccine effective against norovirus infection.
A previous epidemiological study suggested that vitamin A supplementation decreases norovirus infection rates and clinical symptoms3. Moreover, the responses of various intestinal cytokines were modified by vitamin A supplementation during norovirus infection4. Retinoic acid (RA), the metabolite of dietary vitamin A, contributes to both innate and adaptive immune responses5. Additional evidence also indicates that RA deficiency impairs immunity, whereas RA excess can induce inflammatory disorders5. Retinoic acid-inducible gene 1 (RIG-1) and melanoma differentiation-associated gene 5 (MDA5) signaling play crucial roles in antiviral responses to viral RNA by producing type I interferons (IFNs)6. Moreover, a recent study reported that sufficient vitamin A supplementation reduced both mortality and morbidity associated with infectious gastrointestinal and respiratory diseases7.
The gut microbiota plays a pivotal role in pathogen infection and mucosal immune responses through cross-talk with mucosal immune systems8,9. For example, an altered gut microbiome in mice lacking Toll-like receptors (TLRs) and myeloid differentiation primary response gene 88 (Myd88) was strongly associated with metabolic syndrome, type 1 diabetes (T1D) and host defense against microbial infection10,11,12,13. Above all, dietary foods and drugs play a crucial role in modulating gut microbiome diversity, with changes that are directly linked to health conditions. For example, recent studies suggested that Akkermansia muciniphila, which increased significantly in the gut environment as a result of metformin treatment, may improve metabolic diseases such as type 2 diabetes14,15,16.
In this study, we evaluated: 1) compositional changes in the gut microbiota and host immune responses following RA treatment, and 2) the anti-norovirus effects of specific gut microbiota whose abundance was increased by RA treatment (Lactobacillus spp.).
Results
Effect of vitamin A on norovirus replication in vitro and in vivo
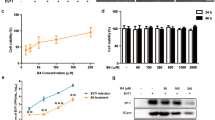
To confirm whether vitamin A inhibits norovirus replication, we first tested the inhibitory effect of retinol on MNV replication in murine RAW 264.7 cells. Retinol treatment (10, 30 and 50 U/mL) significantly inhibited MNV plaque formation at 24, 48 and 72 h (Fig. 1a). A similar inhibitory effect was observed for human norovirus genome replication by monitoring its effect on human norovirus replicon-bearing cells at 24, 48 and 72 h following retinol treatment (Fig. 1b). Twenty-four hours after retinol treatment, the copy number of the human norovirus genome had decreased significantly in the presence of 100 U/ml retinol compared to the negative control. At 48 and 72 h, retinol concentrations above 30 U/mL exhibited a significant inhibitory effect. The effect of RA was assessed based on the experimental infection of ICR mice with MNV, which induced a subclinical infection but no weight loss. Based on these infection conditions, ~60% of MNV-infected mice had cleared MNV at 72 h post-infection, indicating that MNV could establish a persistent infection in approximately 40% of infected mice without RA treatment. In contrast, the rate of MNV persistence in MNV-infected mice decreased to 28.6% in mice treated with 1 mg/kg/day RA.
(a) Inhibition of MNV replication by retinol treatment. MNV (MOI 0.01) was infected into RAW 264.7 cells. Cells were then treated with retinol, and MNV replication was measured using a plaque assay. (b) Expression of the human norovirus genome. HG23 cells were incubated with various concentrations of retinol for 24–72 h. Total RNA of the human norovirus genome was quantified by real-time RT-PCR. In vitro tests were repeated three times. Significance was analyzed by the Mann–Whitney U-test and compared to the negative control. *p < 0.05. Retinol did not exhibit cytotoxicity under the conditions used in this experiment.
Analysis of the gut microbiome
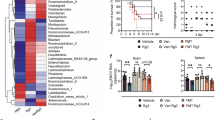
To understand the presumed perturbation in the gut microbiome by either RA administration or MNV infection, a total of 2,670,123 sequences were generated after quality filtering 29 mouse cecum samples. Collected sequences were subsequently analyzed and classified into the following 13 prokaryotic phyla: Actinobacteria, Bacteroidetes, Cyanobacteria, Deferribacteres, Euryarchaeota, Firmicutes, Fusobacteria, Lentisphaerae, Proteobacteria, SR1, Tenericutes, TM7, and Verrucomicrobia. Figure 2a shows the relative gut microbiome abundance at the family level. RA treatment and MNV administration resulted in significantly different bacterial communities (Fig. 2b).
RA administration (RA) (n = 7) and MNV inoculation (n = 5) significantly altered the composition of the gut microbiome compared to the negative control (n = 5). In particular, MNV inoculation significantly altered the composition of the gut microbiota during RA administration (n = 7), and significantly changed bacterial abundances in cecum samples. (a) Relative bacterial abundance was classified at the family level. (b) UniFrac distance in groups categorized by RA administration and MNV inoculation. Samples from the MNV-inoculated group following RA administration showed the most significant differences in bacterial community composition. Statistically significant differences between groups were determined by the Mann–Whitney U-test. *p < 0.05.
Characteristics of the gut microbiome following RA administration
Seven samples from the RA-administered group were clustered in principle coordinate analysis (PCoA) of weighted UniFrac distances (Fig. 3a). Bacterial Allobaculum, Aggregatibacter, Bifidobacterium, Dialister, and Enhydrobacter genera were significantly increased in mice treated with RA (Fig. 4a). Moreover, KEGG pathways predicted by PICRUSt did not show clear clustering by RA administration or bacterial beta diversity (Fig. 3b).
(a) Beta diversity in groups categorized by RA administration and MNV inoculation was assessed by weighted and unweighted principle coordinate analysis (PCoA). Five groups categorized by RA administration and MNV inoculation were clearly clustered in the weighted, but not the unweighted, analysis. This result indicated that the abundances of certain bacterial taxa were changed by MNV inoculation following RA administration. (b) PCoA of KEGG pathways predicted by PICRUSt. MNV inoculation and RA administration also affected the KEGG pathway categories, as well as the bacterial diversity.
(a) Characterization of bacterial abundance by MNV and RA. Significant differences were identified by LEfSe analysis as a p value <0.05 in both the Kruskal–Wallis test (among classes) and Wilcoxon test (between subclasses). The threshold logarithmic LDA score was 3.0. NC: negative control. RA was suspended in corn oil, and administered orally. (b) Abundance of Lactobacillus by RA administration and MNV infection. Among abundant bacteria from the LEfSe analysis, the abundance of Lactobacillus is shown separately. Bacterial abundances were decreased by MNV infection and increased by RA administration after MNV inoculation.
Characteristics of the gut microbiome following MNV infection
In contrast to the effect of RA administration, MNV infection during RA administration significantly affected the gut microbiome. When MNV was infected during RA administration, seven samples were clearly clustered into groups based on beta diversity of the gut microbiome (Fig. 3a). Microbiome diversity was greater in PCoA analysis of weighted UniFrac than unweighted. Figure 3a shows the relative bacterial abundance following MNV inoculation during RA administration. After MNV inoculation, Lactobacillaceae populations were significantly decreased compared to those in negative control (NC) mice (Fig. 4b). Furthermore, the Lactobacillus genus was significantly increased when MNV was inoculated during RA administration compared to other groups. Clustering into five groups was observed for PCoA data based on functional KEGG pathways (Fig. 3b). In addition, among the predicted KEGG pathways, the counts for glycolysis/gluconeogenesis (carbohydrate metabolism), benzoate degradation (xenobiotic biodegradation and metabolism), ABC transporters (membrane transport), the phosphotransferase system (PTS) (membrane transport), and signal transduction mechanisms (cellular processes and signaling) were significantly altered by MNV inoculation during RA administration compared with those that underwent MNV inoculation only.
Inhibition of MNV replication by Lactobacillus strains in vitro
Four Lactobacillus strains (L. ruminis SPM 1308, L. fermentum KCTC 3112, L. rhamnosus KCTC 18427P, and L. reuteri KCTC 18428P) showed a significant inhibitory effect on MNV replication compared to retinol treatment alone (10−1 to 10−2, Fig. 5).
MNV replication was significantly inhibited by four Lactobacillus strains and retinol (50 U/mL). Lactobacillus (estimated at 1 × 105 CFU by OD) was inoculated into MNV-inoculated RAW 264.7 cells (0.01 MOI) for 24 h in the absence of retinol. All Lactobacillus strains significantly decreased the replication of MNV. LPS (10 ng/mL), used as a control, showed an antiviral effect similar to that of Lactobacillus. After freezing and thawing twice, a plaque assay was performed to quantify MNV. Different superscript letters indicate significant differences (p < 0.05) according to Duncan’s post hoc test.
Effect of vitamin A and Lactobacillus on RIG-1 and cytokine expression in vitro
Retinol treatment without MNV infection decreased the expression of RIG-1 in RAW 264.7 cells. In contrast, retinol treatment together with MNV infection significantly increased RIG-1 expression compared to both RIG-1 treatment only and the negative control (Fig. 6a). Moreover, MNV infection significantly increased the expression level of IFN-β, which was further enhanced by retinol treatment (Fig. 6b).
(a) RIG-1 and IFN-β expression in RAW 264.7 cells was measured 24 h after retinol and MNV treatment. (b) The effect of Lactobacillus strains on RIG-1, IFN-β, IFN-γ, and TNF-α expression in MNV-infected RAW264.7 cells was determined 24 h after Lactobacillus treatment. Gene expression was analyzed by SYBR-based quantitative real-time PCR. Different superscript letters indicate significant differences (p < 0.05) according to Duncan’s post hoc test.
RIG-1 expression in MNV-infected RAW 264.7 cells was significantly decreased by L. ruminis and L. fermentum (Fig. 6b). IFN-β and IFN-γ expression was also significantly increased by the four Lactobacillus strains (Fig. 6b). TNF-α expression was not altered significantly by the Lactobacillus strains. LPS showed an antiviral effect with Lactobacillus strains did not increase RIG-1, IFN-β, IFN-γ, and TNF-α expression (Fig. 6b).
Discussion
We demonstrated the inhibitory effect of vitamin A against MNV replication both in vitro and in vivo. Interestingly, these inhibitory effects occurred directly or indirectly via microbiome changes, particularly on Lactobacillus strains in the gut. The prevention of norovirus infection through dietary supplementation has been considered important, as no effective treatment or vaccine against norovirus infection is currently available. This study provides important knowledge regarding the role of particular microorganisms in the gut, such as Lactobacillus spp., in preventing acute viral gastroenteritis.
Specific activation of immune responses by RA may induce favorable gut microbiota changes for protection from MNV infection, although a direct effect of RA on gut microbiota composition was not identified. In previous studies, the gut microbiome was determined to be a key modulator for metabolic disorders in TLR knockout mice, including TLR2 and TLR511,17, and TLR5-mediated sensing of gut microbiota impacted antibody response to influenza vaccination18. Therefore, specific gut microbiota may mediate activation of immune responses by RA to inhibit MNV infection. A previous study reported that MDA5 and TLR3, viral pattern recognition receptors, play distinct roles in the immune response against MNV infection19. Moreover, RIG-1 and MDA5 activation is crucial for the immune response to produce IFNs5,20. Long et al. reported that specific chemokines, including monocyte chemoattractant protein-1 (MCP-1), also referred to as chemokine (C-C motif) ligand 2 (CCL2), and interleukin 8 (IL-8), were involved in the immune response against norovirus infection by RA4. In addition, retinoid X receptor α (RXRα) plays a key role in innate immunity through the upregulation of chemokines Ccl6 and Ccl921. Therefore, these chemokines may mediate antiviral signaling by RA, from viral recognition to inhibition of norovirus.
Interestingly, the gut bacterial community was slightly altered by RA administration, but was significantly altered by MNV inoculation during RA administration. Notably, Lactobacillus spp. levels were remarkably increased by MNV inoculation during RA administration. In previous studies, Lactobacillus spp. exhibited antiviral effects and alleviated the symptoms caused by rotavirus and influenza viral infections22,23. Moreover, it has been reported that probiotics, including Lactobacillus spp., exert various beneficial effects on human health through their immunomodulatory activities24,25. In this study, we demonstrated that Lactobacillus spp. was enriched by vitamin A intake and significantly inhibited MNV replication.
Over the past several decades, different Lactobacillus strains have been isolated from food and human stool and subsequently provided to individuals as probiotics for health purposes26,27. In this study, we describe Lactobacillus strains with antiviral effects, as observed from artificial changes in the gut microbiota. Recent studies demonstrated that gut microbiota was not only influenced by xenobiotics metabolism, but also by medications. Specifically, exposure to certain medications yielded changes in gut microbiota, which resulted in beneficial effects on metabolic disorders14,16,28. Likewise, vitamin A clearly exerted antiviral effects via modulation of gut microbiota, such as Lactobacillus spp.
IFN-β plays a key role in the antiviral response mediated by the RIG-1/MDA5 pathway5,20. It has been reported that certain Lactobacillus strains activate IFN-β through induction of RIG-129,30. In this study, Lactobacillus strains significantly upregulated IFN-β and IFN-γ expression in MNV-infected RAW 264.7 cells. In a recent study, IFN-γ was reported to play an important role in adaptive immunity to MNV infection31. Unlike retinol, Lactobacillus did not increase the expression of RIG-1, while L. ruminis and L. fermentum slightly decreased RIG-1 expression in MNV-infected RAW 264.7 cells. Therefore, both IFN-β and IFN-γ are involved in the anti-MNV immune response, in a manner not mediated by the RIG-1/MDA5 pathway. In addition, LPS, which exerts an antiviral effect by inducing the production of various cytokines including IFNs, inhibited the replication of MNV in this study32,33, but did not increase IFN expression, unlike Lactobacillus.
MNV infection was recently shown not to cause major changes in the intestinal microbiota of Swiss Webster and inbred C57BL/6 mice34. In contrast, the present study showed that MNV inoculation caused a significant alteration in the gut microbiome; the abundance of Proteobacteria was increased by MNV inoculation, consistent with observations in patients whose gut microbiota was perturbed by human norovirus infection35. Moreover, in patients with abundant Proteobacteria, the abundance of Alcaligenaceae was less than in healthy controls35. In the in vivo study described herein, the decreased abundance of Lactobacillaceae, Alcaligenaceae and Veillonellaceae revealed another significant change in the gut microbiota following MNV infection. Therefore, we believe that the abundance of these bacteria may serve as potential diagnostic and preventive markers for human norovirus infection after specificity issues are addressed.
Most importantly, focus should be placed on norovirus genotypes, which based on previous studies, could be a critical factor in changes in the microbiota, immune responses and viral susceptibility. Norovirus susceptibility in mice is highly associated with MNV genogroups in mouse infection models using six MNV genotypes36. Expression profiles of cytokines differ among norovirus genogroups4. Moreover, a previous epidemiological study reported that the effect of RA varied depending on the norovirus genotype3. Therefore, the gut microbiota composition specific to pathogenic or clinical norovirus infections should be further investigated.
Due to issues with conventional in vitro cell culture and the mouse model of human norovirus, MNV has been widely used as a surrogate in pathological and inhibitory studies37,38. In several recent studies including ours, gut microbiome changes by norovirus were not coincident34,35. MNV infectivity and pathology differed according to the mouse strain used19,39. The different results in terms of the effect of MNV on the gut microbiome in our study compared to previous studies was likely due to use of a different mouse model34. Disruption of the gut microbiome by human norovirus reported in a previous human study suggested that the gut microbiome is closely related to the immune response to norovirus35. The ICR mouse model with MNV-1 used herein may not be the ideal mouse model to study human norovirus. However, further studies will provide insight into the role of the gut microbiome in norovirus infection.
In conclusion, we demonstrated the inhibitory effect of vitamin A on MNV both in vitro and in vivo, which had only been reported in a human epidemiological study to date. In addition, specific gut microbiota (e.g., Lactobacillus spp.) were significantly altered by MNV inoculation during RA administration, and Lactobacillus spp. only significantly inhibited MNV replication in vitro independent of retinol treatment. We also show that IFN-β is involved in the antiviral immune response. In the future, we expect that the specific modulation of gut microbiota could have an impact on determining functional bacteria for both preventive and medicinal uses.
Methods
Cells, viruses and reagents
Huh-7-based NV replicon-bearing cells (HG23 cells) were kindly provided by Dr. Chang40. RAW 264.7 cells were maintained in Dulbecco’s Minimal Essential Medium (DMEM) containing 10% fetal bovine serum (Gibco), 10 mM HEPES, 10 mM sodium bicarbonate, 10 mM nonessential amino acids, and 50 μg/mL gentamicin. Retinol (95144; Sigma-Aldrich) was applied to cells following dilution with ethanol. Prior to in vitro experiments with retinol, cell viability was confirmed by propidium iodide (PI) staining using a FACScan system (FACSCalibur; BD Biosciences). Prior to all in vitro experiments, cell viability against retinol was test using EZ-CyTox Cell Viability Assay kit (Daeil Lab Service Co., Ltd, Korea).
In vitro assay to examine MNV inhibition by retinol
Retinol (10, 30, 50, 70, and 100 U/mL) was applied to 80–90% confluent HG23 cells in a 6-well plate. Human norovirus replicon expression was analyzed 24, 48 and 72 h after treatment. Total RNA was extracted using the easy-spin Total RNA Extraction Kit (iNtRON) according to the manufacturer’s instructions. The reaction mixture comprised the AgPath-ID One-Step RT-PCR kit (Ambion). Norovirus GI-specific primers and FAM-labeled GI probes for amplification were described previously41. Amplification was performed with a 7300 Real-time PCR System (Applied Biosystems) under the following conditions: reverse transcription (RT) for 10 min at 42 °C, initial denaturation at 95 °C for 10 min, and 45 cycles of amplification, with denaturation at 95 °C for 15 s and annealing plus extension at 60 °C for 60 s. A standard curve was generated using serial dilutions of a norovirus GI-type clone, as described previously42. Moreover, inhibition of MNV replication was tested in RAW 264.7 cells. Retinol (10, 30 and 50 U/mL) was applied to 0.01 MOI MNV-inoculated RAW 264.7 cells. MNV replication was analyzed by a plaque assay as described previously43 and fluorescence activated cell sorting (FACS) at 24, 48 and 72 h after inoculation.
Animal model
Twelve-week-old male ICR mice were purchased from KOATECH. (Pyeongtaek, South Korea) and housed in an animal biosafety level 2 (ABL-2) facility at Seoul National University College of Medicine, South Korea. RA (R-2625; Sigma-Aldrich), suspended in corn oil, was administered orally to mice at a dosage of 1 mg/kg/day for eight days. On the seventh day, mice were infected perorally with 5 × 106 plaque forming units (PFUs) of MNV-1 suspended in phosphate-buffered saline (PBS) by oral gavage. The spleen was removed for MNV quantification, homogenized using a PT-2000 E homogenizer (PT-2000 E; POLYTRON) and centrifuged at 10,000 × g for 30 min. The resulting supernatants were used for MNV detection.
All experimental protocols in this study were approved by the Seoul National University Institutional Animal Care and Use Committee and were conducted in accordance with the Guide for the Care and Use of Laboratory Animal (SNU-111208-4).
Analysis of the gut microbiome with and without vitamin A and MNV infection
The gut microbiome was analyzed from mouse whole cecum samples including fecal materials. Cecum samples were homogenized using a homogenizer (PT-2000 E; POLYTRON) and by bead beating with a PowerBead Tube (MO BIO Laboratories Inc.). Total DNA was extracted using the QIAamp® DNA Stool Mini Kit (Qiagen) and QIAcube system (Qiagen). Partial sequences of 16S rRNA genes were amplified according to the 16S rRNA amplification protocol from the Earth Microbiome Project44. For each sample, 16S rRNA genes were amplified using the 515F/806R primer set for amplification of the V4 region (515F forward primer: 5′-AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGT GTGCCAGCMGCCGCGGTAA -3′; 806R reverse primer containing a unique 12-base barcode for tagging each PCR product: 5′-CAAGCAGAAGACGGCATACGAGAT- NNNNNNNNNNNN -AGTCAGTCAGCC GGACTACHVGGGTWTCTAAT -3′). Amplified polymerase chain reaction (PCR) products were purified using the UltraClean® PCR Clean-Up Kit (MO BIO Laboratories Inc.). Sequencing of partial bacterial 16S rRNA genes was performed using the MiSeq Reagent Kit V3 (600 cycles) and MiSeq platform (Illumina).
Prior to analysis of the 16S rRNA sequences, BCL files were converted into raw FASTQ files including read1, index and read2 sequences using CASAVA-1.8.2. After preprocessing (quality filtering and trimming steps using FASTX-Toolkit), sequences were assigned to operational taxonomic units (OTUs, 97% identity), and representative sequences were selected using QIIME 1.7.0 software45. Next, taxonomic composition, alpha diversity and beta diversity were analyzed. LDA Effect Size (LEfSe) was used to estimate taxonomic abundance and characterize differences between groups46. In addition, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict functional genes in the sampled microbial community based on the KEGG pathway database47. A heat map of functional gene abundance was generated using MultiExperiment Viewer (MEV) software (v4.8.1).
In vitro assay to examine MNV inhibition by retinol and Lactobacillus
The Lactobacillus strain L. ruminis SPM 1308 was obtained from Dr. Nam-Joo Ha at Sahmyook University. L. fermentum KCTC 3112 was obtained from Korean Collection for Type Cultures (KCTC). L. rhamnosus KCTC 18427P and L. reuteri KCTC 18428P were additionally isolated in fresh feces of Korean infant. Prior to the inhibition assay, Lactobacillus quantity was estimated using optical density (OD). A cell viability test was performed and the concentration of the antibiotic gentamycin (100 μg/mL) was determined, which represents the steady state of Lactobacillus. MNV (MOI 0.01) was infected into RAW 264.7 cells for 1 h. Following a media change, Lactobacillus (estimated to 1 × 105 CFU by OD), retinol (50 U/mL), and LPS (10 ng/mL) were inoculated for 24 h in MNV-inoculated RAW 264.7 cells. After 24 h, cells were performed a plaque assay to quantify MNV after freezing and thawing twice.
RNA extraction and gene expression analysis by quantitative real-time PCR in vitro
Expression levels of RIG-1, IFN-β, IFN-γ, and TNF-α were analyzed at 24 h after retinol, MNV, and Lactobacillus treatment in RAW 264.7 cells. Total RNA was extracted using the easy-spin™ Total RNA Extraction Kit (iNtRON). Then cDNA was synthesized using the High Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer’s instructions. To estimate the expression levels of cytokine mRNAs, a QuantiTect® SYBR® Green PCR Kit (204143; Qiagen) and a 7300 Real Time PCR System (Applied Biosystems) were used. The reaction mixture (25 μL) for real-time PCR was composed of 2× QuantiTect SYBR Green PCR Master Mix (12.5 μL), primers (RIG-1: forward CAAAAACCACCCATACAATCAG and reverse CAAATGTGATGTGTACAGGAAG, IFN-β: forward ATAAGCAGCTCCAGCTCCAAG and reverse GTCTCATTCCACCCAGTGCTG, IFN-γ: forward TGAACGCTACACACTGCATC and reverse CGACTCCTTTTCCGCTTCCT, and TNF-α: forward GCCACCACGCTCTTCTGCCT and reverse GGCTGATGGTGTGGGTGAGG, each 50 pmol in 0.2 μL), RNase-free water (11.1 μL), and template DNA (1 μL). GAPDH was used as the internal control.
Statistical analysis
All features are expressed as the mean ± standard deviation (SD) of each group. Significant differences in the UniFrac distance between groups were analyzed by the Mann–Whitney U-test. In a relative abundance analysis using LEfSe based on the Kruskal–Wallis and Wilcoxon tests, significance was defined as a p value <0.05. The threshold on the logarithmic LDA score was set as 2.0–3.0. To quantity the level of mRNA in vivo relative to that of an internal control (GAPDH), the 2−ΔΔCt relative quantification method (ΔΔCt = (Ct.Target − Ct.GAPDH)Group1 − (Ct.Target − Ct.GAPDH)Group2) was used. Statistical significance was assessed by one-way ANOVA followed by Duncan’s post hoc test. All statistical analyses were performed using SPSS software ver. 12.0. A p value <0.05 was considered to indicate statistical significance.
Additional Information
How to cite this article: Lee, H. and Ko, G. P. Antiviral effect of vitamin A on norovirus infection via modulation of the gut microbiome. Sci. Rep. 6, 25835; doi: 10.1038/srep25835 (2016).
References
Ahmed, S. M., Lopman, B. A. & Levy, K. A systematic review and meta-analysis of the global seasonality of norovirus. PloS one 8, e75922, doi: 10.1371/journal.pone.0075922 (2013).
Hall, A. J. et al. Epidemiology of foodborne norovirus outbreaks, United States, 2001–2008. Emerg Infect Dis 18, 1566–1573, doi: 10.3201/eid1810.120833 (2012).
Long KZ1 et al. Vitamin A supplementation has divergent effects on norovirus infections and clinical symptoms among Mexican children. The Journal of infectious diseases 196, 978–985, doi: 10.1086/521195 (2007).
Long, K. Z. et al. Vitamin A modifies the intestinal chemokine and cytokine responses to norovirus infection in Mexican children. J Nutr 141, 957–963, doi: 10.3945/jn.110.132134 (2011).
Hall, J. A., Grainger, J. R., Spencer, S. P. & Belkaid, Y. The role of retinoic acid in tolerance and immunity. Immunity 35, 13–22, doi: 10.1016/j.immuni.2011.07.002 (2011).
Takeuchi, O. & Akira, S. Innate immunity to virus infection. Immunol Rev 227, 75–86, doi: 10.1111/j.1600-065X.2008.00737.x (2009).
Thornton, K. A., Mora-Plazas, M., Marin, C. & Villamor, E. Vitamin A deficiency is associated with gastrointestinal and respiratory morbidity in school-age children. J Nutr 144, 496–503, doi: 10.3945/jn.113.185876 (2014).
Kamada, N., Seo, S. U., Chen, G. Y. & Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nature reviews. Immunology 13, 321–335, doi: 10.1038/nri3430 (2013).
Chu, H. & Mazmanian, S. K. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 14, 668–675, doi: 10.1038/ni.2635 (2013).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241, doi: 10.1016/j.cell.2004.07.002 (2004).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231, doi: 10.1126/science.1179721 (2010).
Backhed, F. Host responses to the human microbiome. Nutr Rev 70 Suppl 1, S14–17, doi: 10.1111/j.1753-4887.2012.00496.x (2012).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113, doi: 10.1038/nature07336 (2008).
Lee, H. & Ko, G. Effect of metformin on metabolic improvement and gut microbiota. Applied and environmental microbiology 80, 5935–5943, doi: 10.1128/AEM.01357-14 (2014).
Shin, N. R. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut, doi: 10.1136/gutjnl-2012-303839 (2013).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 110, 9066–9071, doi: 10.1073/pnas.1219451110 (2013).
Caricilli, A. M. et al. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS biology 9, e1001212, doi: 10.1371/journal.pbio.1001212 (2011).
Oh, J. Z. et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41, 478–492, doi: 10.1016/j.immuni.2014.08.009 (2014).
McCartney, S. A. et al. MDA-5 recognition of a murine norovirus. PLoS Pathog 4, e1000108, doi: 10.1371/journal.ppat.1000108 (2008).
Broquet, A. H., Hirata, Y., McAllister, C. S. & Kagnoff, M. F. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. Journal of immunology 186, 1618–1626, doi: 10.4049/jimmunol.1002862 (2011).
Nunez, V. et al. Retinoid X receptor alpha controls innate inflammatory responses through the up-regulation of chemokine expression. Proc Natl Acad Sci USA 107, 10626–10631, doi: 10.1073/pnas.0913545107 (2010).
Sindhu, K. N. et al. Immune Response and Intestinal Permeability in Children With Acute Gastroenteritis Treated With Lactobacillus rhamnosus GG: A Randomized, Double-Blind, Placebo-Controlled Trial. Clin Infect Dis 58, 1107–1115, doi: 10.1093/cid/ciu065 (2014).
Kikuchi, Y. et al. Oral Administration of Lactobacillus plantarum Strain AYA Enhances IgA Secretion and Provides Survival Protection against Influenza Virus Infection in Mice. PloS one 9, e86416, doi: 10.1371/journal.pone.0086416 (2014).
Tsai, Y. T., Cheng, P. C. & Pan, T. M. The immunomodulatory effects of lactic acid bacteria for improving immune functions and benefits. Appl Microbiol Biotechnol 96, 853–862, doi: 10.1007/s00253-012-4407-3 (2012).
Sanders, M. E. Impact of probiotics on colonizing microbiota of the gut. J Clin Gastroenterol 45 Suppl, S115–119, doi: 10.1097/MCG.0b013e318227414a (2011).
Swain, M. R., Anandharaj, M., Ray, R. C. & Parveen Rani, R. Fermented fruits and vegetables of Asia: a potential source of probiotics. Biotechnology research international 2014, 250424, doi: 10.1155/2014/250424 (2014).
de Vrese, M. & Schrezenmeir, J. Probiotics, prebiotics, and synbiotics. Advances in biochemical engineering/biotechnology 111, 1–66, doi: 10.1007/10_2008_097 (2008).
Haiser, H. J. & Turnbaugh, P. J. Developing a metagenomic view of xenobiotic metabolism. Pharmacological research : the official journal of the Italian Pharmacological Society 69, 21–31, doi: 10.1016/j.phrs.2012.07.009 (2013).
Tomosada, Y. et al. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC immunology 14, 40, doi: 10.1186/1471-2172-14-40 (2013).
Miettinen, M. et al. Nonpathogenic Lactobacillus rhamnosus activates the inflammasome and antiviral responses in human macrophages. Gut microbes 3, 510–522, doi: 10.4161/gmic.21736 (2012).
Nice, T. J. et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273, doi: 10.1126/science.1258100 (2015).
Karta, M. R. et al. LPS modulates rhinovirus-induced chemokine secretion in monocytes and macrophages. American journal of respiratory cell and molecular biology 51, 125–134, doi: 10.1165/rcmb.2013-0404OC (2014).
Simard, S., Maurais, E., Gilbert, C. & Tremblay, M. J. LPS reduces HIV-1 replication in primary human macrophages partly through an endogenous production of type I interferons. Clinical immunology 127, 198–205, doi: 10.1016/j.clim.2008.01.007 (2008).
Adam, M. Nelson et al. Murine norovirus infection does not cause major disruptions in the murine intestinal microbiota. Microbiome 1, doi: 10.1186/2049-2618-1-7 (2013).
Nelson, A. M. et al. Disruption of the human gut microbiota following Norovirus infection. PloS one 7, e48224, doi: 10.1371/journal.pone.0048224 (2012).
Thackray, L. B. et al. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. Journal of virology 81, 10460–10473, doi: 10.1128/JVI.00783-07 (2007).
Wobus, C. E., Thackray, L. B. & Virgin, H. W. t. Murine norovirus: a model system to study norovirus biology and pathogenesis. Journal of virology 80, 5104–5112, doi: 10.1128/JVI.02346-05 (2006).
Hirneisen, K. A. & Kniel, K. E. Comparing human norovirus surrogates: murine norovirus and Tulane virus. J Food Prot 76, 139–143, doi: 10.4315/0362-028X.JFP-12-216 (2013).
Ward, J. M. et al. Pathology of immunodeficient mice with naturally occurring murine norovirus infection. Toxicol Pathol 34, 708–715, doi: 10.1080/01926230600918876 (2006).
Chang, K. O., Sosnovtsev, S. V., Belliot, G., King, A. D. & Green, K. Y. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 353, 463–473, doi: S0042-6822(06)00396-5 (2006).
Kageyama, T. et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol 41, 1548–1557, doi: 10.1128/JCM.41.4.1548–1557.2003 (2003).
Park, Y., Cho, Y. H., Jee, Y. & Ko, G. Immunomagnetic separation combined with real-time reverse transcriptase PCR assays for detection of norovirus in contaminated food. Appl Environ Microbiol 74, 4226–4230, doi: AEM.00013-08 (2008).
Lee, J., Zoh, K. & Ko, G. Inactivation and UV disinfection of murine norovirus with TiO2 under various environmental conditions. Applied and environmental microbiology 74, 2111–2117, doi: 10.1128/AEM.02442-07 (2008).
Gilbert, J. A. et al. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand Genomic Sci 3, 243–248, doi: 10.4056/sigs.1433550 (2010).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, doi: 10.1038/nmeth.f.303 (2010).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60, doi: 10.1186/gb-2011-12-6-r60 (2011).
Langille, M. G. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821, doi: 10.1038/nbt.2676 (2013).
Acknowledgements
This work was supported by the National Research Foundation of Korea NRF-2015R1A2A1A10054078.
Author information
Authors and Affiliations
Contributions
H.L. and G.P.K. designed the study. H.L. performed all experiments and data analysis. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lee, H., Ko, G. Antiviral effect of vitamin A on norovirus infection via modulation of the gut microbiome. Sci Rep 6, 25835 (2016). https://doi.org/10.1038/srep25835
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep25835
This article is cited by
-
All-trans retinoic acid alleviates transmissible gastroenteritis virus-induced intestinal inflammation and barrier dysfunction in weaned piglets
Journal of Animal Science and Biotechnology (2024)
-
Chemokine CCL6 Plays Key Role in the Inhibitory Effect of Vitamin A on Norovirus Infection
Journal of Microbiology (2023)
-
All-trans retinoic acid increases the pathogenicity of the H9N2 influenza virus in mice
Virology Journal (2022)
-
Probiotics protect against RSV infection by modulating the microbiota-alveolar-macrophage axis
Acta Pharmacologica Sinica (2021)
-
The Interaction Between Viruses and Intestinal Microbiota: A Review
Current Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.